Difference between revisions of "EEGLAB"
| (42 intermediate revisions by 3 users not shown) | |||
| Line 1: | Line 1: | ||
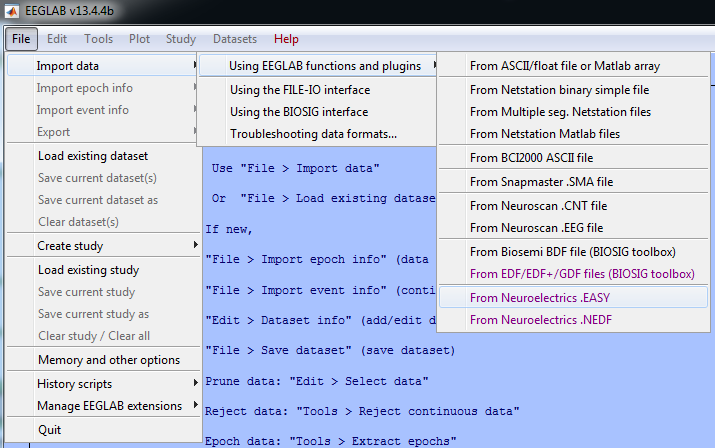
| − | [[File: | + | [[File:ImportFiles.png|600px|thumb|right|Import EASY and NEDF files with EEGLAB toolbox]] |
[http://sccn.ucsd.edu/eeglab/ '''EEGLAB'''] is a powerful MATLAB toolbox used by researchers to process and analyse EEG data. EEGLAB is an interactive platform that includes independent component analysis (ICA), time/frequency analysis, artifact rejection, event-related statistics, and several useful modes of visualization of the averaged and single-trial data. For further information about the EELAB toolbox capabilities, check the [http://sccn.ucsd.edu/wiki/EEGLAB '''EEGLAB wiki webpage''']. | [http://sccn.ucsd.edu/eeglab/ '''EEGLAB'''] is a powerful MATLAB toolbox used by researchers to process and analyse EEG data. EEGLAB is an interactive platform that includes independent component analysis (ICA), time/frequency analysis, artifact rejection, event-related statistics, and several useful modes of visualization of the averaged and single-trial data. For further information about the EELAB toolbox capabilities, check the [http://sccn.ucsd.edu/wiki/EEGLAB '''EEGLAB wiki webpage''']. | ||
| − | Due to its popularity among the neuroscience community, Neuroelectrics has developed an EEGLAB plugin to make it compatible with the files generated by NIC - Neuroelectrics Instrument Controller software. The NE EEGLAB NIC Plugin allows you to directly load NIC output files: plain text files (.easy) and binary NIC files (.nedf). The NE EEGLAB NIC Plugin can be downloaded for free from the [http://www.neuroelectrics.com/ | + | Due to its popularity among the neuroscience community, Neuroelectrics has developed an EEGLAB plugin to make it compatible with the files generated by NIC - Neuroelectrics Instrument Controller software. The NE EEGLAB NIC Plugin allows you to directly load NIC output files: plain text files (.easy) and binary NIC files (.nedf). The NE EEGLAB NIC Plugin can be downloaded for free from the [http://www.neuroelectrics.com/downloads/ '''Downloads'''] of the website. |
| − | Download now the EEGLAB plugin and start processing and analysing your EEG data. Using EEGLAB with NIC files is now easier than ever! | + | Download now the EEGLAB plugin, add it in the plugins folder of EEGLAB and start processing and analysing your EEG data. Using EEGLAB with NIC files is now easier than ever! |
| + | |||
| + | |||
| + | The EEGLAB Plugin for NIC files, not only allows you to upload the NIC output files onto the MATLAB, but it also includes the geometric coordinates of the electrode positions for the Enobio 20 and Enobio 32 models. Please note that the coordinates of the extension channel of Enobio 20 need to be assigned manually according to the position chosen. | ||
| + | |||
| + | For Enobio or Starstim 20 and 32 standard cap, polar and cartesian electrode positions can be found [[media:Electrode_Positions.zip |'''here''']]. | ||
| + | |||
| + | The EEGLAB NIC Plugin by default has a Location folder with location files. These files should be modified according to the channel location of the experiment. | ||
| + | |||
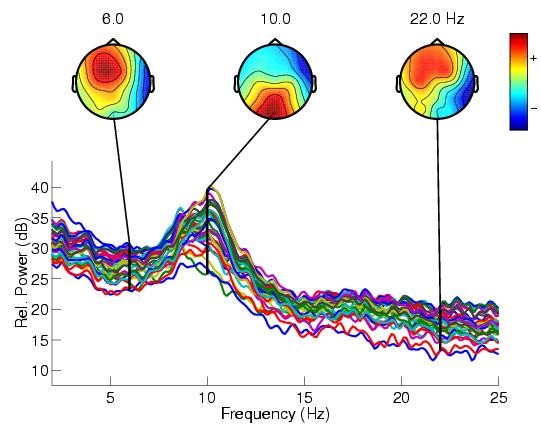
| + | [[File:eeglab3.jpeg|1000px|thumb|left|Power Spectrum and Scalp Maps generated by EEGLAB toolbox]] | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | == Tutorial Outline == | ||
| + | |||
| + | [ftp://sccn.ucsd.edu/pub/eeglabtutorial5.00b.pdf This tutorial] will demonstrate (hands-on) how to use EEGLAB to interactively preprocess, analyze, and visualize the dynamics of event-related EEG, MEG, or other electrophysiological data by operating on the tutorial EEG dataset "eeglab_data.set" which you may download [http://sccn.ucsd.edu/eeglab/download/eeglab_data.set here (4Mb)]. With this dataset, you should be able to reproduce the sample actions discussed in the tutorial and get the same (or equivalent) results as shown in the many results figures. | ||
Latest revision as of 16:02, 22 January 2019
EEGLAB is a powerful MATLAB toolbox used by researchers to process and analyse EEG data. EEGLAB is an interactive platform that includes independent component analysis (ICA), time/frequency analysis, artifact rejection, event-related statistics, and several useful modes of visualization of the averaged and single-trial data. For further information about the EELAB toolbox capabilities, check the EEGLAB wiki webpage.
Due to its popularity among the neuroscience community, Neuroelectrics has developed an EEGLAB plugin to make it compatible with the files generated by NIC - Neuroelectrics Instrument Controller software. The NE EEGLAB NIC Plugin allows you to directly load NIC output files: plain text files (.easy) and binary NIC files (.nedf). The NE EEGLAB NIC Plugin can be downloaded for free from the Downloads of the website. Download now the EEGLAB plugin, add it in the plugins folder of EEGLAB and start processing and analysing your EEG data. Using EEGLAB with NIC files is now easier than ever!
The EEGLAB Plugin for NIC files, not only allows you to upload the NIC output files onto the MATLAB, but it also includes the geometric coordinates of the electrode positions for the Enobio 20 and Enobio 32 models. Please note that the coordinates of the extension channel of Enobio 20 need to be assigned manually according to the position chosen.
For Enobio or Starstim 20 and 32 standard cap, polar and cartesian electrode positions can be found here.
The EEGLAB NIC Plugin by default has a Location folder with location files. These files should be modified according to the channel location of the experiment.
Tutorial Outline
This tutorial will demonstrate (hands-on) how to use EEGLAB to interactively preprocess, analyze, and visualize the dynamics of event-related EEG, MEG, or other electrophysiological data by operating on the tutorial EEG dataset "eeglab_data.set" which you may download here (4Mb). With this dataset, you should be able to reproduce the sample actions discussed in the tutorial and get the same (or equivalent) results as shown in the many results figures.